Introduction
The presence of clonally expanded mutant hematopoietic stem cells, clonal hematopoiesis (CH) is known to predispose to hematologic malignancy, particularly myeloid neoplasms (MN). However, the overall risk of developing MNs among individuals with CH is low (<1% per year). Limitations in our ability to predict who with CH will progress to MN is a barrier to prevention strategies. The human proteome reflects the downstream impact of complex interactions between genetic and epigenetic regulation of biologic systems. We hypothesized that plasma proteomic markers might be associated with MN risk and if so could inform our understanding of the mechanisms promoting the progression of CH to MN.
Methods
Among 46,237 individuals in the UK Biobank (UKB) with whole exome sequencing data and single nucleotide polymorphism (SNP) array profiling of the blood and plasma proteomic profiling using the Olink Explore platform (1,463 unique proteins), we jointly characterized CH and plasma proteomic profile. Cox proportional hazard models were used to assess the association between the level of each plasma protein and risk of MN; while controlling for age, sex, genetic ancestry, smoking status, blood counts, and sample collection year. The relative contribution of clinical, CH, and proteomics features in prediction of incident MN was assessed using lasso regression with 100-fold cross validation. We performed protein quantitative trait loci (pQTL) mapping to identify independent germline autosomal genetic loci that significantly regulate plasma proteomic levels (p <5x10 -8). We calculated genetically predicted risk scores for plasma protein levels as the as the sum of the effect estimates from linear regression analysis with additive genetic effect, multiplied by the number of risk alleles for each locus. As a validation cohort, we used an independent set of 427,614 individuals in the UKB to test for an association between genetically predicted levels of proteomic markers and MN risk.
Results
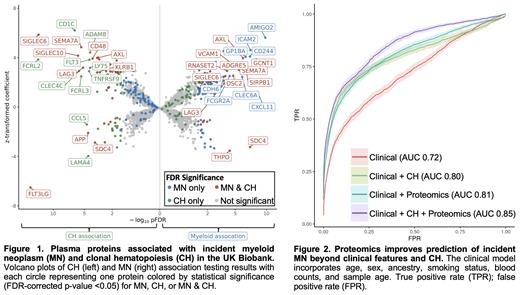
A total of 115 participants developed MN at least 3 months after study entry (median 83 months). As expected, participants who developed MN (cases) had a higher frequency of CH at the time of study entry compared to those free of hematologic malignancy (controls); 42 versus 6% for SNV/Indels and 23 versus 3% for mosaic copy number events in cases compared to controls respectively. We identified 115 proteins significantly associated (qFDR <0.05) with incident MN (Figure 1). These include known regulators of the adaptive immune system, innate immune system, leukocyte trafficking, hematopoietic stem cell self-renewal, and platelet activation.
Given that CH is a genetic precursor to MN, we assessed the relationship between plasma proteomic markers of MN and CH. Among the 115 plasma proteins that were significantly associated with MN, 35 were significantly (qFDR <0.05) and 59 nominally (p <0.05) associated with CH. After adjusting for CH, the strength of the association between plasma protein markers and MN risk was generally reduced (mean 7%), but remained significant for many (N=44). We identified a total of 79 plasma proteins that were associated with the presence of CH at the time of study entry, 34 of which overlapped with MN risk proteins. Inclusion of proteomics features in a lasso regression model (AUC = 0.85) improved prediction of incident MN beyond clinical factors and CH (AUC = 0.80; Figure 2).
There were 1,282 independent genetic loci that were significantly associated with 115 proteomic markers of MN risk. On average, genetic risk scores captured 12% of the total variance of plasma protein levels (range 0.1-64%). In an independent cohort of 427,614 UKB participants (1,099 of whom developed MN), we found that of the 110 plasma protein markers with a genetic risk score R 2 >1%, 12 were significantly associated with MN (p <0.05). Of the remaining, 63 were directionally consistent.
Conclusions
We show that plasma proteomic markers predict risk of MN. Some, but not all of the impact of plasma proteomic profile on MN risk is mediated by baseline CH. Inclusion of proteomic markers improves MN risk prediction beyond clinical and CH features. Genetic risk score analysis provides orthogonal validation of the plasma proteomic signature for MN risk. These data highlight the promise of protein and genetic biomarker integration for early diagnosis of MN and further support the role of immune cell regulation in early leukemogenesis.
Disclosures
Oh:CTI BioPharma, Bristol Myers Squibb, Disc Medicine, Blueprint Medicines, PharmaEssentia, Constellation/MorphoSys, Geron, AbbVie, Sierra Oncology/GSK, Cogent, Incyte, Morphic, Protagonist: Consultancy. Link:Roche: Other: Roche provided Idasanutlin free of charge. No compensation was provided, and Roche was not involved in design, conduction, or analysis of experiments.. Bolton:Servier: Research Funding; GoodCell: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal